Technology Development Project 4
Principal investigators: Curtis Langlotz, Michael Tilkin, Maryellen Giger, and Paul Kinahan
Collaborative development of common user access portal, technology infrastructure and data management processes across MIDC organizations and technology stacks.
Updated January 20, 2023
Access by researchers to MIDRC is enabled through a MIDRC Data Commons Portal. Data intake and distribution leverages existing technology, networks, and infrastructure that span the members of the consortium, as well as investigators throughout the nation.
Immediate data processing and distribution during startup phase.
the Cancer Imaging Archive and Fred Prior (University of Arkansas)
In the first 3 months while the data infrastructure for MIDRC was being integrated under a data commons, public facing data intake, processing, storage, and dissemination was provided through the Cancer Imaging Archive (TCIA). TCIA has extensive experience curating and hosting public-facing imaging data.
This project has now completed.
Data ingestion from RSNA and accessible through the MIDRC Portal.
George Shih, MD (Cornell University)
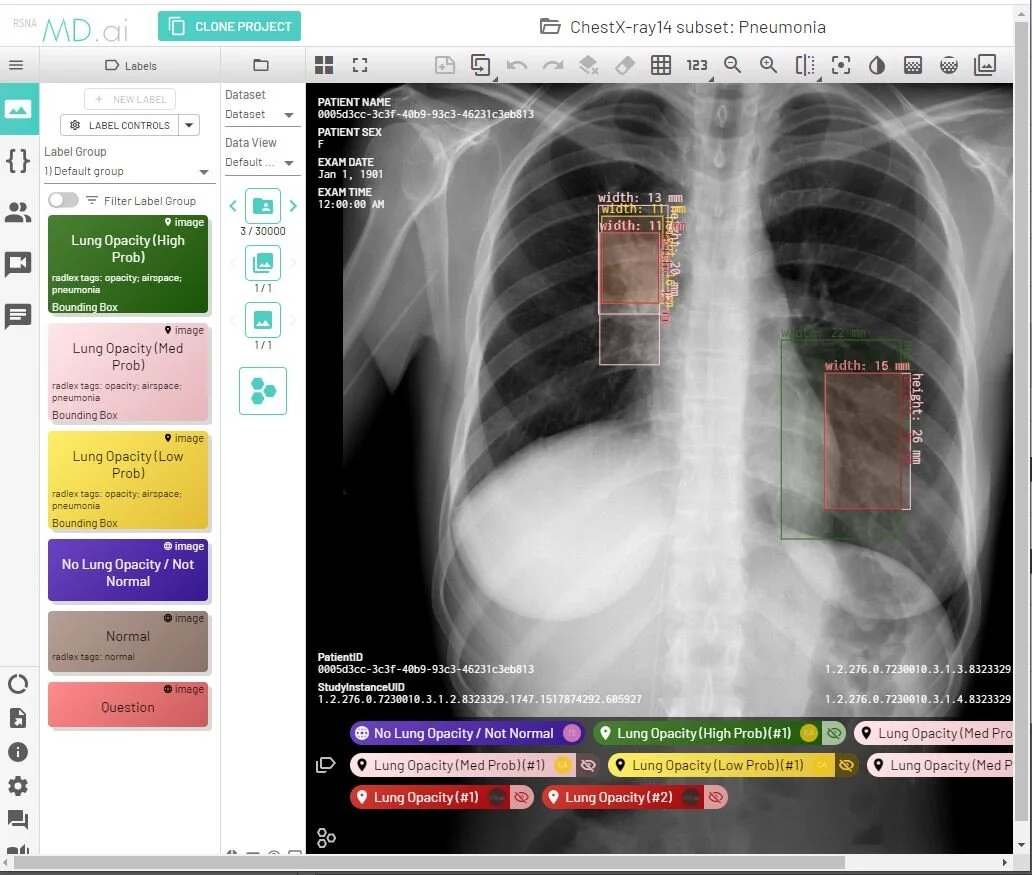
The RSNA enables rapid batch ingestion of large volumes of COVID-19 imaging and associated data according to the common data model and processes developed under MIDRC. RSNA has been collaborating in this effort with the ACR and other data intake organizations, which are working together on harmonized methods to process and validate data ingested by MIDRC. This cloud-native infrastructure is being integrated with the MD.ai image labeling platform amongst others.
Example annotation of a MIDRC chest X-ray
Development of common data intake, processing, quality control, and distribution processes across intake organizations.
Paul Kinahan (University of Washington), Michael Tilkin (ACR), and Curtis Langlotz (Stanford)
Data intake leverages existing technology, networks, and infrastructure that span the members of the consortium. The common data intake process leverages real-time contributions from healthcare organizations, batched contributions from healthcare organizations, and batched patient data contributions, as well as in kind efforts by industrial partners, as available.
Ingestion from ACR TRIAD and accessible through the MIDRC Portal.
Michael Tilkin (ACR) and Laura Coombs (ACR)
The ACR has leveraged and extended the TRIAD network and DART data toolkit to support the goals of the open MIDRC discovery platform. ACR TRIAD is an image exchange platform that is the standard for the NCI National Clinical Trial Network. The DART data platform provides an archive for both clinical and image data, a user access portal for search and analysis, image processing pipelines for machine learning and advanced analytics, and the ability to download research data sets.
Creation of an MIDRC Data Commons Portal based on a common data model and user experience.
Robert Grossman (University of Chicago), Curtis Langlotz (Stanford), Michael Tilkin (ACR), and Maryellen Giger (University of Chicago)
The ongoing aims of this project are to provide a seamless data exploration and analysis experience for MIDRC users, and to provide common authentication and object identification services with other NIH data commons. An instance of the Gen3 Data Platform has been deployed, which is an open source (with an Apache License) software for developing and operating cloud-based data commons. The Gen3 platform also provides workspaces, and an ecosystem of cloud-based applications and notebooks to create data ecosystems. MIDRC employs the Gen3 Data Commons, which uses a graph data model to store clinical, phenotype and other structured information.
Currently, these TDP4 sub-projects are tasked to
Extend existing data dictionary and data collection models to address Long-COVID and additional imaging modalities typically associated with cardiac and neurological system involvement
Extend harmonization of deidentification methods to address new imaging modalities associated with cardiovascular and neurological systems
Implement across data intake pathways the harmonized approach recommended under TDP 2a for de-identification of head images.
Adapt ACR and RSNA data curation pipeline to incorporate LOINC mapping as defined by DQH to generate and submit common study descriptions.
Leverage RSNA’s & ACR’s imaging expertise to contribute to annotations workgroup
Continue to evolve the data model to reflect long COVID population data, including appropriate clinical data for identification of long COVID patients and incorporation of neuro and ultrasound images.
Continue to improve the automation and QC process for data ingestion, including supporting data packages, the data ingestion required for the DICOM viewer, and related needs.
Continue to evolve the functionality of the MIDRC data portal
Continue to evolve the functionality of the DICOM viewer
Integrate and/or develop a MIDRC annotation system
Improve the automation around data sequestration and support for challenges
Support execution through standard interfaces of submitted containerized pipelines for data analysis and machine learning to support both remote and federated machine learning and data analysis
Continue to update and evolve the data ingestion pipelines and processes as required, to support Long-COVID and other MIDRC requirements.
Extend contributing site relationships to include Long-COVID imaging studies and clinical data and usage of data for new research project objectives. Continue to update and evolve the data ingestion pipelines and processes as required, to support Long-COVID and other MIDRC requirements.