Technology Development Project 3
Principal investigators: Maryellen Giger and Paul Kinahan
Develop and implement quality assurance and evaluation procedures for usage across MIDRC.
Updated January 20, 2023
MIDRC’s initiative spans from data ingestion through data access and dissemination, including data quality metrics, data provenance, data processing audits paths, and task-directed data distribution. Use of the platform also provides opportunity to initiate discovery of yet unknown trends and develop new tools to predict diagnosis and outcomes at an individual patient and population level. MIDRC uses data management methods harmonized across all participating organizations at three critical stages: (1) intake, including curation, de-identification, abstraction, and quality assessment (2) annotation and labelling of imaging and other data using semi-automated approaches and (3) distributed access and query methods. These methods have yielded, and continue to yield, a large hybrid and distributed data set that is in accordance with the FAIR principles (findable, accessible, interoperable and reusable).
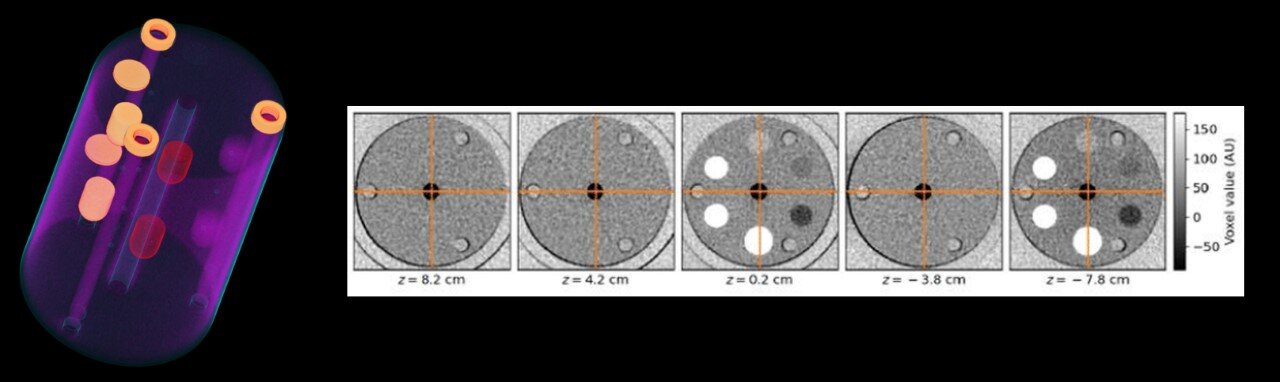
Development of digital and physical imaging phantoms for COVID data.
Paul Kinahan (University of Washington) and John Boone (University of California-Davis)
Digital and physical phantoms are needed both to test individual components of the data stream and to check results throughout the overall data stream. We have conducted research to determine the appropriate digital and physical imaging phantoms for COVID data at each step. This project has used pre-existing phantom data (e.g. ACR CT phantoms) collected by contributing sites by working with AAPM members that are diagnostic medical physicists at selected sites. It has also determined whether a new phantom design that is more specific for COVID-19 patient images allows for more accurate or efficient measurement of imaging quality. Learn more about TDP3a…
Assessment of image quality on ingestion into MIDRC.
John Boone (University of California-Davis) and Paul Kinahan (University of Washington)
A wide variety of imaging systems and protocols is used to collect the images provided to MIDRC. The issue of data heterogeneity is well-known, but this project provides an opportunity to perform and track a consistent and harmonized assessment of the quality of the intake data. A key component of this project has been to determine the image quality metrics described above both before and after the image de-identification and normalization steps illustrated by the technology stacks. This is beingused to determine if image information is being modified in a manner that could impact downstream analysis by MIDRC users. This assessment may also be applied on image output from MIDRC as a quality control measure.
This project is adding experts in MRI to handle the increased complexities of DICOM meta-information for long Covid, and add the needed data structures for 'long COVID' studies that use multiple modalities, starting initially with MRI, XR, and CT.
Development of benchmarking methods for the various technology assessment and clinical tasks in COVID-19 research and translation.
Michael McNitt-Gray (University of California-Los Angeles), Berkman Sahiner (FDA), and Karen Drukker (University of Chicago)
Appropriate performance metrics for machine learning algorithms are being determined and benchmarking of performance has been performed. A decision tree tool has been developed to recommend appropriate performance metrics for different machine learning tasks. The purpose of this decision tree tool is to give advice to researchers on how to efficiently and effectively evaluate performance of their AI methods which will ultimately help translate these new AI methods to clinical practice. Benchmarking of performance will incorporate sequestered datasets from MIDRC that are drawn from specific distributions.
This project will extend its scope for long COVID, i.e., to develop benchmarking methods for multi-modality tasks (extending CXR and CT to MRI and others), for decision making task that are affected by analyses of the brain and cardiac images, and, especially, for temporal tasks that require assessment of metrics over time for prognosis, response, post-COVID risks. Learn more about TDP3c…
Development of task-based distribution methods.
Kyle Myers (Puente Solutions LLC), Heather Whitney (University of Chicago), Natalie Baughan (University of Chicago), and Maryellen Giger (University of Chicago)
To facilitate FDA clearance and accelerate clinical usage, methods of selecting clinical cases for independent testing are required to appropriately evaluate a ‘mature’ algorithm deemed ready for clinical translation. In this project, the construct of task-based distribution for independent testing is being developed. Methodology has been developed to draw samples, i.e., distributions of cases, for a specific clinical task, (e.g., diagnosis of COVID-19 from other presentations of pneumonia) matched to the clinical claim and intended patient population, to be randomly drawn from the larger sequestered dataset within the MIDRC (generalizable to any dataset), and subsequently used in the testing.
This project is being extended for long COVID, i.e., to incorporate temporal-based data elements into the sequestering process for the handling of multiple exams over time, additional imaging modalities, and additional anatomical regions (beyond thorax). Likewise, methods for task-based distribution sampling will be extended to AI claims on prognosis, response, and risk over time.